More efficient data retrieval from synthetic polymer data storage
Increasing amounts of data require storage, often for long
periods. Synthetic polymers are an alternative to conventional storage
media because they maintain stored information while using less space
and energy. However, data retrieval by mass spectrometry limits the
length and thus the storage capacity of individual polymer chains. In
the journal Angewandte Chemie, researchers have now introduced a
method that overcomes this limitation and allows direct access to
specific bits without reading the entire chain.
© Wiley-VCH, re-use with credit to 'Angewandte Chemie' and a link to the original article.
Data accumulates constantly, resulting from business
transactions, process monitoring, quality assurance, or tracking product
batches. Archiving this data for decades requires much space and energy.
For long-term archival of large amounts of data that requires infrequent
access, macromolecules with a defined sequence, like DNA and synthetic
polymers, are an attractive alternative.
Synthetic polymers have advantages over DNA: simple
synthesis, higher storage density, and stability under harsh conditions.
Their disadvantage is that the information encoded in polymers is
decoded by mass spectrometry (MS) or tandem-mass sequencing (MS2).
For these methods, the size of the molecules must be limited, which
severely limits the storage capacity of each polymer chain. In addition,
the complete chain must be decoded in sequence, building block by
building block—the bits of interest cannot be accessed directly. It is
like having to read through an entire book instead of just opening it to
the relevant page. In contrast, long chains of DNA can be cut into
fragments of random length, sequenced individually, and then
computationally reconstructed into the original sequence.
Kyoung Taek Kim and his team at the Department of Chemistry
at Seoul National University (Rep. Korea) have developed a new method by
which very long synthetic polymer chains whose molecular weights greatly
exceed the analytical limits of MS and MS2 can be efficiently
decoded. As a demonstration, the team encoded their university address
into ASCII and translated this—together with an error detection code
(CRC, an established method used to ensure data integrity)—into a binary
code, a sequence of ones and zeroes. This 512-bit sequence was stored in
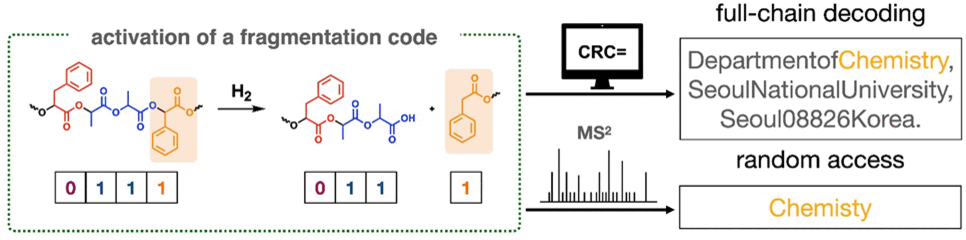
a polymer chain made of two different monomers: lactic acid to represent
a 1 and phenyllactic acid to represent a 0. At irregular intervals, they
also included fragmentation codes containing mandelic acid. When
chemically activated, the chains break at those locations. In their
demonstration, they obtained 18 fragments of various sizes that could be
individually decoded by MS2 sequencing.
Specially developed software initially identified the
fragments based on their mass and their end groups, as shown by the MS
spectra. During the MS2 process, previously measured
molecular ions brake down further, and these pieces were then also
analyzed. The fragments could be sequenced based on the mass difference
of the pieces. With the aid of the CRC error detection code, the
software reconstructed the sequence of the entire chain, overcoming the
length limit for the polymer chains.
The team was also able to decode interesting bits without
sequencing the entire polymer chain (random access), such as the word
“chemistry” in the code for their address. By taking into account that
the parts of their address are all in a specific order (department,
institution, city, postal code, country) and separated by commas they
were able to isolate the location where the desired information was
stored within the chain and only sequenced the relevant fragments.
(3686 characters)
About the Author
Dr Kyoung Taek Kim is a Professor of Polymer Chemistry at
Seoul National University. His research interest is to relate the
chemical structures of polymers to their structures and functions via
the development of efficient methods for precision polymers and block
copolymers, including sequence-defined polymers and self-replicating
polymers.
Copy free of charge—we would appreciate a transcript/link of your
article. The original articles that our press releases are based on can
be found in our online pressroom.